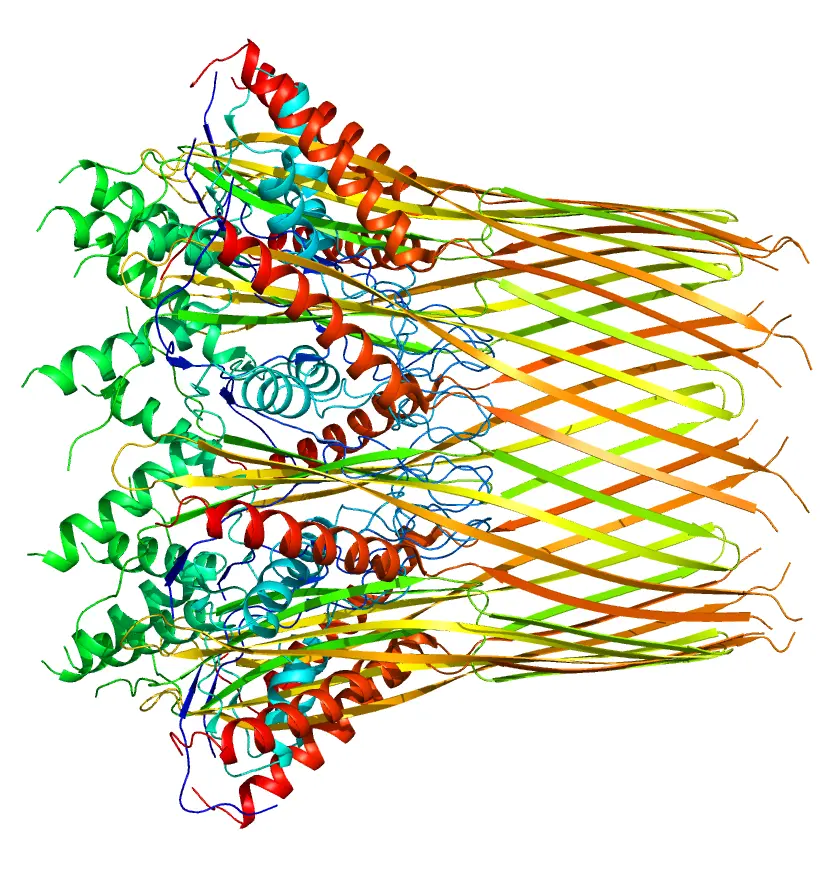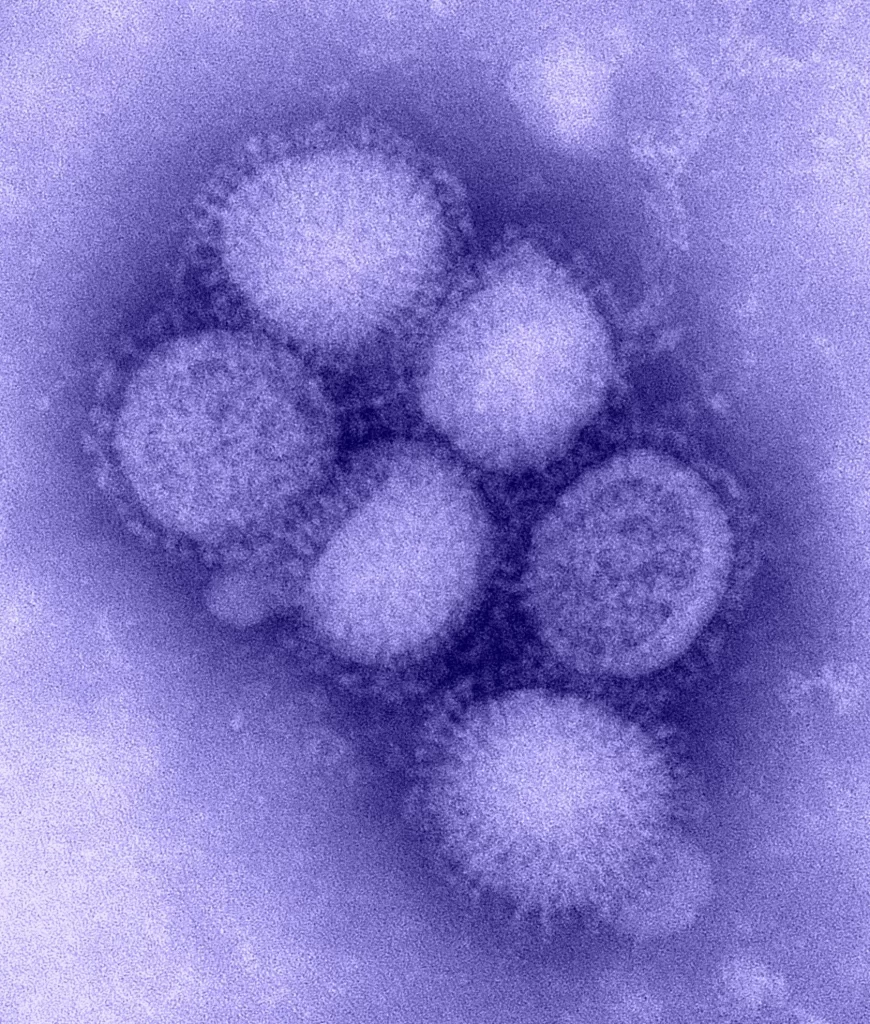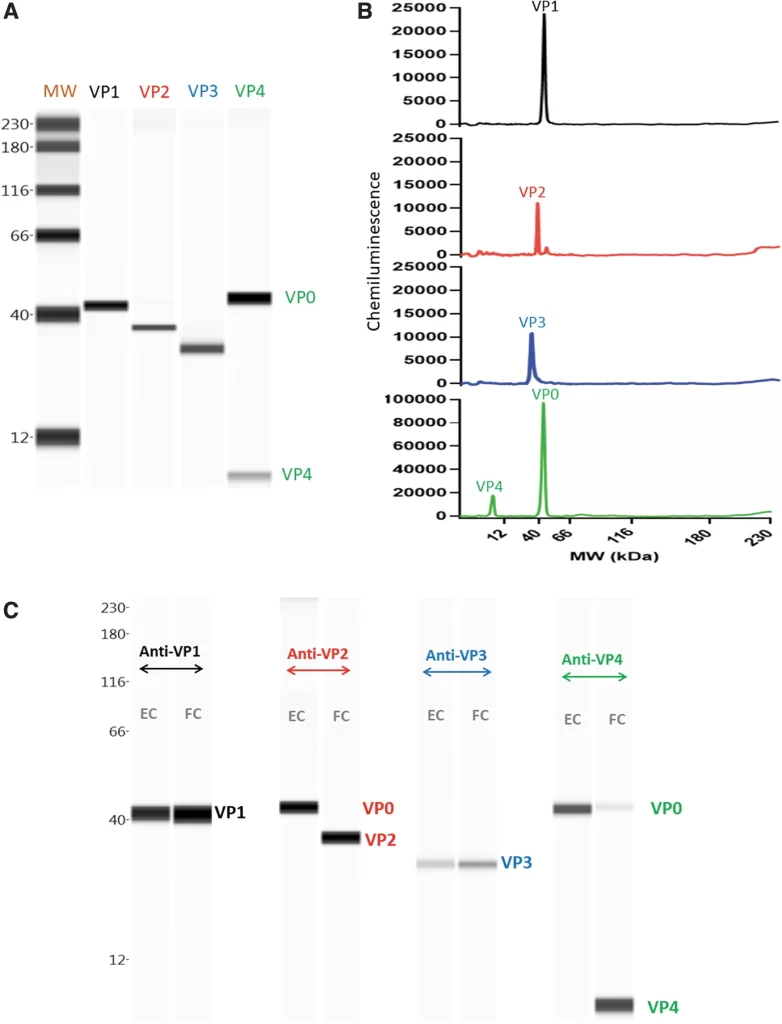
Oncolytic viruses are a novel type of virus that specifically targets and eliminates tumor cells, and have been an emerging immunotherapy agent. One such is Coxsackievirus A21 (CVA21), which has been demonstrating a healthy safety profile as well as desired targeting of the cancerous cells. While producing drugs that can properly utilize this virus, there rises a need to characterize the ratio of total particles to empty ones, thus researchers developed a method to see Coxsackievirus A21 viral proteins quantified with capillary Western and peptides made up of the components of the capsid proteins.
CVA21 characterized with capillary Western and capsid peptides
LifeTein provided the group with the capsid peptides, VP1, VP2, VP3, and VP4, as well as the antibodies necessary for the testing. Results utilizing these peptides and antibodies included total particle concentration, empty particle concentration, as well as the empty to full ratio. Being able to characterize this kind of information in this downstream process will prove to be immensely valuable in future studies.
The Simple Western method displayed by the researchers proved itself to be an invaluable tool for optimizing decisions for these types of oncolytic virus studies. Consistency in these tests will surely lead to massive amounts of time being saved in future drug development and assessments. LifeTein will always be ready to lend a hand to any work needing peptides or antibodies to further their research in the broad field of drug application.
Paul F. Gillespie, Richard R. Rustandi, Andrew R. Swartz, Liang Shang, Jessica Raffaele, Ashley Prout, Nicholas Cunningham, Mohamed Dawod, James Z. Deng, Shiyi Wang, Jessica Olson, Yvonne Shieh, and John W. Loughney.
Quantitation of Coxsackievirus A21 Viral Proteins in Mixtures of Empty and Full Capsids Using Capillary Western.
Human Gene Therapy.Jan 2023.68-77.http://doi.org/10.1089/hum.2022.147