There are a large number of protein-protein interactions in the cell. These interactions regulate all fundamental cellular processes such as cell cycle and signal transduction. In this summary, we selected some methods to identify and analyze protein-protein interactions.
Introducing: Peptide Conjugation Magnetic Agarose, 10% bead suspension for pull-down experiment, peptide-protein binding, protein-protein interaction, and immunoprecipitation.
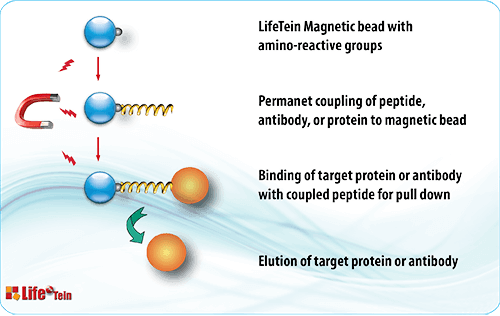
The most straightforward method is to use the peptide as bait in affinity pull-down experiments and followed by detection of binding proteins. The synthetic peptides are used to verify the protein-protein interactions by competitive disruption of binding.
For example, biotinylated peptides and photocleavable biotin peptides can be immobilized onto avidin-conjugated sepharose under controlled conditions. Peptides with a free cysteine can be cross-linked to nanoparticles, resins, or sepharoses to make affinity column. Peptides can be tagged with 6His, FLAG, V5 HA, or cMYC for protein affinity chromatography.
The protocol used to Pull-Down Biotin-labeled Peptides.
Immobilized streptavidin beads were mixed with biotinylated peptide before incubation with cell lysates. Cells were lysed in 1% (v/v) Nonidet P-40 buffer with protease and phosphatase inhibitors. Proteins were incubated with the immobilized peptides at 4 °C for 6 h. After washes, eluted bound proteins by boiling in SDS buffer. Eluates from the active and control bait peptide pull-downs were used for further analysis.
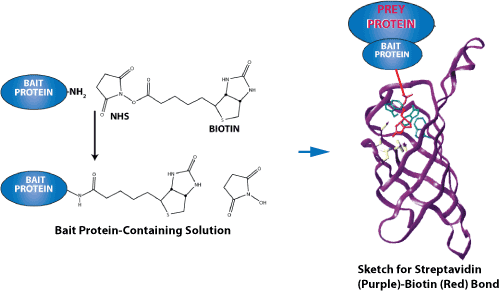
Sketch for streptavidin-biotin binding. Streptavidin-biotin and avidin-biotin are the most prominent ligand-receptor pairs.
Reference:
Acetylation of TUG Protein Promotes the Accumulation of GLUT4 Glucose Transporters in an Insulin-Responsive Intracellular Compartment, The Journal of Biological Chemistry, January 5, 2015, 10.1074/jbc.M114.603977
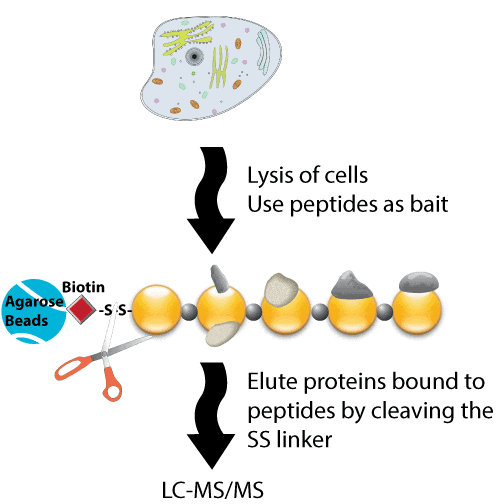
LifeTein synthesized multiple peptides with biotin on the N terminus with an aminohexanoic acid linker. Multiple lysines were acetylated in the synthetic peptides. These peptides were bound to streptavidin-agarose beads and used for pulldown experiments using cell lysates. The peptide acetylation is a valuable tool to study molecular regulation and interactions.
Proteins can be fractionated by SDS-PAGE, and identified by their ability to bind a peptide, protein, or ligand.
One of the most commonly used methods for verification of protein-protein interactions is Co-immunoprecipitation (Co-IP). The first step is to generate the cell lysates. Then an antibody is added. The antigen is precipitated and washed. The final bound complexes are analyzed.
LifeTein's peptide antibody service uses peptide affinity purification steps to remove non-specific antibodies. The antisera are usually made by coupling of the peptide to the KLH carrier protein. Our peptide affinity purified antibodies remove antibody against the carrier KLH protein.
Usually, highly specific antibodies are required for Co-IP. However, an antibody directed against the tag (instead of against the bait protein) can then be used if cells transfected with a plasmid encoding a tagged bait protein. The epitope-tagged proteins can often be eluted by incubation with competing peptides. This specific elution often reduces the amount of contaminating proteins in the eluate.
Performing a coimmunoprecipitating reaction and identifying physiological protein-protein interactions have many advantages. The proteins are at their natural state of posttranslational modification, so the protein-protein interactions are genuine. However, the coimmunoprecipitating can be difficult and generate significant background. This is because of the nature of the indirect interactions, nonspecific binding and antibody contamination that may mask detection. It is therefore important to conduct parallel negative controls, deliberately add an excess of the antigen, or covalently crosslink the proteins before immunoprecipitation.
Co-IP Optimization
Membrane-permeable cross-linking reagents are used in vivo. The excess amino groups can inhibit cross-linking. In addition, the cross-linked protein can be released from the immune complex by cleavage of the crosslink with DTT. After the cross-linking, immunoprecipitation of the ligand protein is performed. Peptides or proteins can be labeled before cross-linking. This method will give a clean result.
LifeTein routinely synthesizes biotinylated peptides for use in protein-protein interaction studies. Although biotin can be introduced either N- or C-terminally (via lysine residues), we recommend the use of an N-terminal modification owing to its low cost, higher success rate, shorter turnaround time, and ease of operation. Peptides are synthesized from the C-terminus to the N-terminus; therefore, the N-terminal modification is the last step in the SPPS protocol, and no additional specific coupling steps are required. In contrast, a C-terminal modification requires additional steps, and is usually more complex. However, in principle the biotin can be positioned anywhere.
Biotin can be separated from the peptide by a variety of different linkers or spacers. Nevertheless, it is recommended that a flexible spacer such as Ahx (a 6-carbon linker) is included to render the biotin label more stable or flexible.
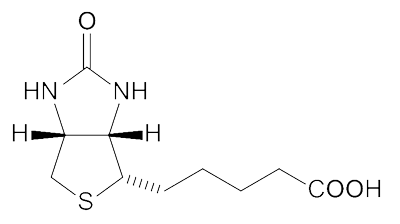
Biotin Structure
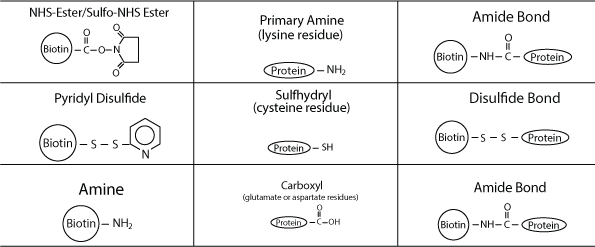
Biotin-labeled peptides are commonly detected or purified using avidin conjugates for many protein-based research applications, including western blotting, ELISA, immunoprecipitation (IP), cell surface labeling, affinity purification, immunohistochemistry (IHC), and flow cytometry/fluorescence-activated cell sorting (FACS).
Biotinylated peptides can be used in conjunction with ELISAs or western blotting with streptavidin-HRP or affinity purified receptors or antibodies. Biotinylated peptides can also be immobilized tightly on streptavidin columns. Nevertheless, an N-terminal amine or an amine group on a lysine residue is needed for successful binding reactions.
Please click here to get a peptide synthesis service quote now!