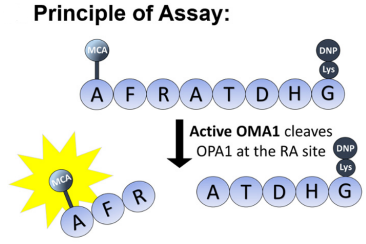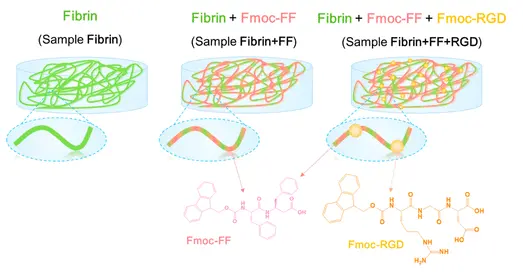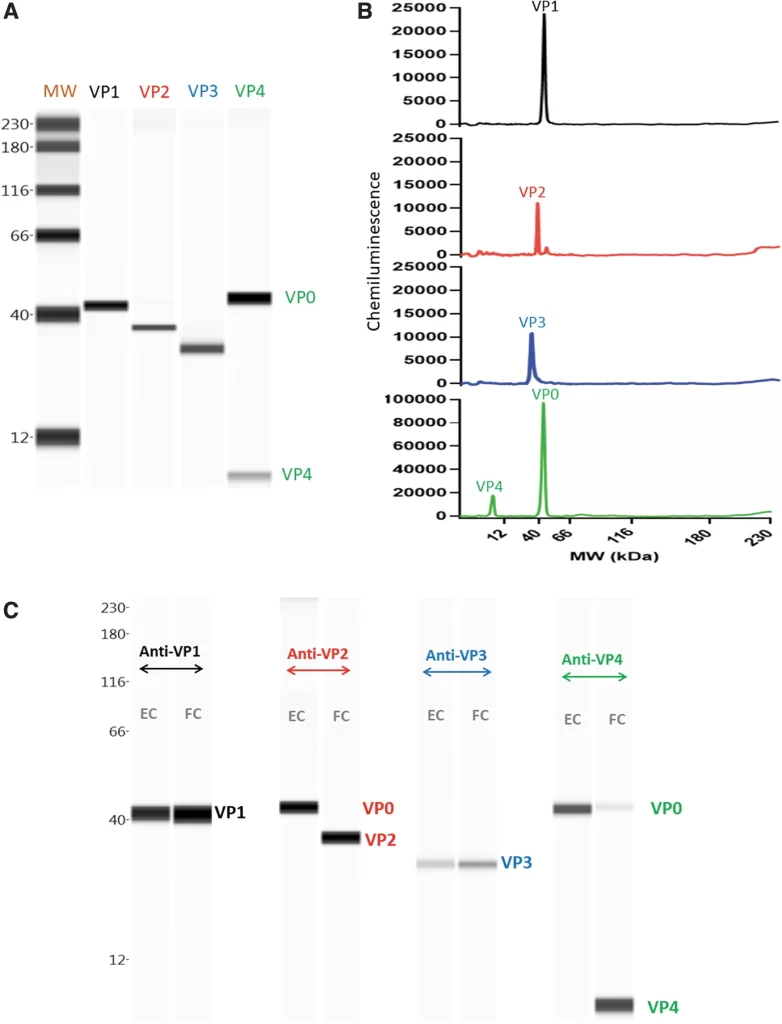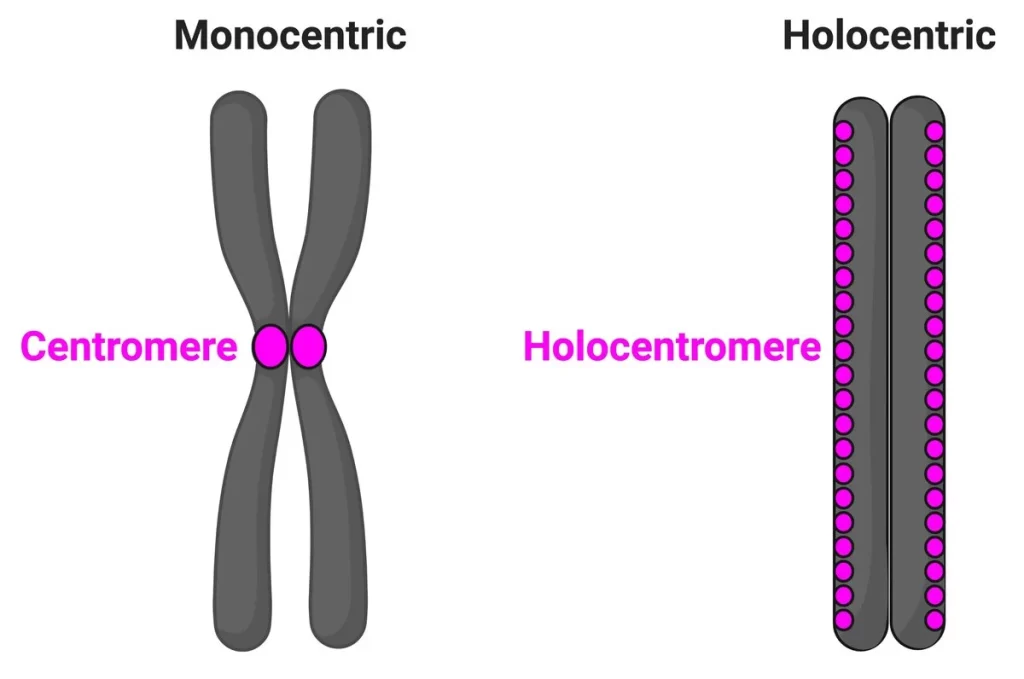
During cell division, microtubules in the chromosome attach to a region called the centromere. While most species have a single size-restricted centromere, or a monocentromere, some species exist with multiple centromeres distributed across the chromosome, called holocentromeres. What is even more interesting is how holocentric chromosomes are considered to have evolved from the monocentric organisms, and this transition occurred independently across distant lineages, such as green algae, protozoans, invertebrates, as well as flowering plant families. One group aimed to study these holocentromeres more via the lilioid Chionographis japonica. Their goal was to better understand the convergent evolution of holocentromeres studied with peptides.
Peptides help explore holocentromeres
The group determined that the chromosomal localization of the target centromere is usually marked with histone H3 (CENH3). With this knowledge, they utilized peptides and antibodies of CENH3 provided by LifeTein to create models of the transition of C. japonica from interphase to prophase and study the possible mechanisms as well. They found the holocentromere was made up only of a few, evenly spaced CENH3-positive megabase-sized satellite arrays. Overall, the reason for the convergent evolution of holocentromeres from a monocentromere may stem from multiple factors, but more experiments like the ones presented will surely provide further analysis into this complex and fascinating case of convergent evolution.
Kuo, YT., Câmara, A.S., Schubert, V. et al. Holocentromeres can consist of merely a few megabase-sized satellite arrays. Nat Commun 14, 3502 (2023). https://doi.org/10.1038/s41467-023-38922-7