When it comes to the world of peptide research, the selection of appropriate fluorescent dyes is crucial for various applications, including cellular imaging, molecular diagnostics, and therapeutic interventions. This article delves into the nuances of choosing the right fluorescent dyes for peptides, offering insights into the latest research and practical considerations.
Key Takeaways:
- The choice of fluorescent dye depends on the specific application and properties of the peptide.
- Commonly used dyes include FITC, FAM, TAMRA, and Cyanine dyes.
- The impact of the dye on the function and location of the peptide should be carefully evaluated.
Understanding Fluorescent Dyes in Peptide Research
The Role of Fluorescent Dyes
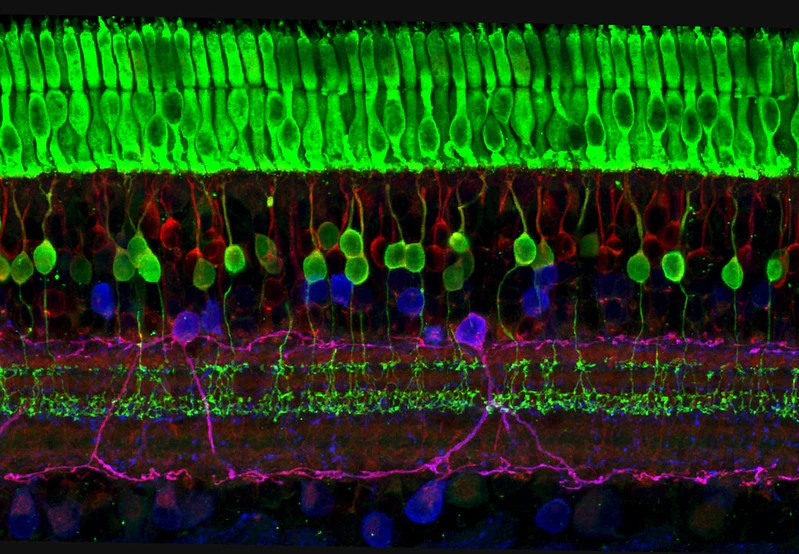
Fluorescent dyes are pivotal in peptide research for visualizing and tracking biological processes. They enable the observation of peptides in various environments, from in vitro studies to in vivo applications.
Selection Criteria
When selecting a fluorescent dye, consider factors like wavelength, brightness, photostability, and the potential impact on peptide structure and function.
Popular Fluorescent Dyes for Peptides
FITC, FAM, TAMRA, and Cyanine Dyes
These dyes are widely used due to their effective labeling properties and compatibility with various imaging techniques. For more information, visit LifeTein’s page on fluorescent dyes.
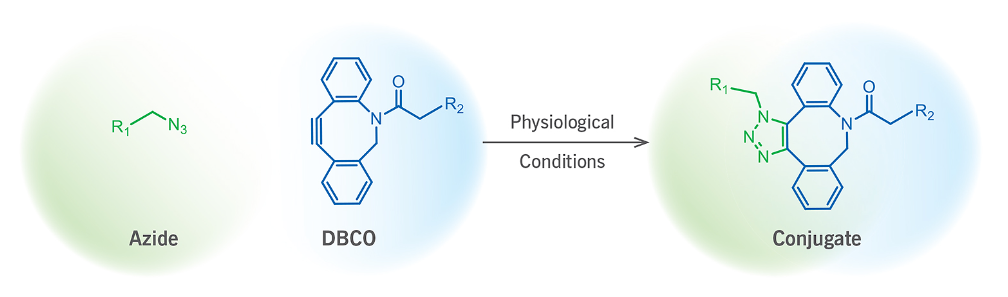
Alexa Dyes are another common option, though these are typically conjugated to peptides via cysteine residues or Lys(N3), like Cyanine dyes.
Novel Dyes and Custom Solutions
Advancements in dye technology have led to the development of novel dyes offering enhanced properties. Custom solutions may also be available for specific research needs.
Impact of Dyes on Peptide Function
Alteration of Peptide Properties
Research indicates that fluorescent labels can significantly alter the physicochemical properties of peptides, affecting their function and localization. For instance, a study by H. Szeto et al. (read more) highlights how different fluorescent labels can lead to varied intracellular targeting and function in cell-penetrating tetrapeptides.
Mitochondrial Targeting and Protection
Certain dyes have been shown to target specific cellular components, such as mitochondria, influencing peptide behavior and therapeutic potential.

Practical Considerations in Dye Selection
Compatibility with Experimental Conditions
The chosen dye must be compatible with the experimental conditions, including pH, temperature, and the presence of other biomolecules.
Cost and Availability
Consider the cost and availability of dyes, especially for large-scale studies or specialized applications. For a range of options, explore LifeTein’s custom synthesis page.
Typically, FITC, FAM, and TAMRA are less costly than dyes like Cyanine or AlexaFluor.
Frequently Asked Questions
- How do I choose the right fluorescent dye for my peptide?
- Consider the application, desired wavelength, required properties of the dye, and the potential impact on the peptide’s function.
- Can the dye alter the function of my peptide?
- Yes, fluorescent labels can change the peptide’s properties and intracellular behavior.
- Are there custom dye options available for specific needs?
- Yes, custom dye solutions can be developed for unique research requirements.
For further reading on the impact of fluorescent dyes on peptides, consider the research by M. Berezin et al. on the selection of small peptide molecular probes (read the study).
Szeto, H.H., Schiller, P.W., Zhao, K. and Luo, G. (2005), Fluorescent dyes alter intracellular targeting and function of cell-penetrating tetrapeptides. The FASEB Journal, 19: 118-120. https://doi.org/10.1096/fj.04-1982fje
Mikhail Y. Berezin, Kevin Guo, Walter Akers, Joseph Livingston, Metasebya Solomon, Hyeran Lee, Kexian Liang, Anthony Agee, and Samuel Achilefu, Rational Approach To Select Small Peptide Molecular Probes Labeled with Fluorescent Cyanine Dyes for in Vivo Optical Imaging. Biochemistry 2011 50 (13), 2691-2700
https://doi.org/10.1021/bi2000966