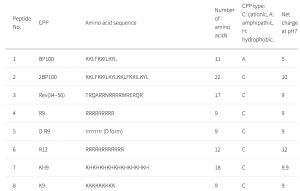
Protein-protein interactions (PPI) are highly specific electrostatic attractions between protein structures. The interactions regulate cell function and influence physiology and development. Mass spectrometry is often used to detect protein-protein interactions. However, all proteomic screens are differential and require two samples such as IP and mock IP, POI and wildtype, or POI and knockout.
The peptide/ biotin-peptide or peptide/fluorescence-labeled peptide are excellent tools for studying protein-protein interactions. Check this link for details: Protein-Protein Interactions: Methods for Detection and Analysis.
https://www.lifetein.com/Peptide_Modifications_biotinylation.html
The compounds that can modulate PPI are hard to discover because the proteins have multiple binding sites and the screening assays are not reliable. In many cases, two or more proteins may interact with one another and form a complex. The optical fluorescence-based methods such as the Cy5, Cy7, FAM, FITC, TAMRA-labeled peptides, or FRET assay are particularly useful in these circumstances. Click for more details: https://www.lifetein.com/Peptide-Synthesis-FITC-modification.html.
The interactions between a fluorescently labeled or intrinsically fluorescent sample and a binding patterner are measured during the application. The changes in intrinsic fluorescence from tryptophan and tyrosine residues in the protein can be measured, which indicates transitions in the protein’s folding state.
The scientists have been working on fusion-based bifunctional proteins in cancer immunotherapy. The bifunctional protein sent an apoptotic signal to the tumor cells and enhanced their killing. The click chemistry is the perfect tool for the drug-protein or protein-protein conjugation. The more we understand the natural receptor-ligand complex and how it might signal, the better we can guide the design of therapeutic agonists. Click here for the peptide conjugation details: https://www.lifetein.com/price_modification_labeling.html